This is the third in a three-part series from Medscape on the impact of AI on drug discovery and development. Part 1 is about AI’s role in designing speedier, more effective clinical trials. Part 2 is about the use of AI to find new applications for existing drugs.
What if we told you that the same technology behind deepfake photos could soon bring new and improved drugs to your patients?
Unlike some of those photos, that’s not fake news.
Researchers are borrowing from artificial intelligence (AI) models, such as OpenAI’s DALL-E, that translate text into images to design never-before-seen proteins from scratch. These designs could form the basis of future drugs or vaccines.

“For the first time, we are seeing AI systems able to generate highly realistic protein structures in a controllable way in seconds,” said Namrata Anand, PhD, a bioengineer and computer scientist.
In 2022, Anand used this technology to develop the model that spawned her company Diffuse Bio. Also that year, a de novo protein designed with AI by researchers at the University of Washington, Seattle, gained clinical approval abroad as a COVID-19 vaccine. Biotech company Absci’s de novo antibody for inflammatory bowel disease is undergoing investigational new drug-enabling studies in hopes of starting clinical trials in 2025. Other AI-enhanced efforts focus on evolving known proteins or finding ways to target proteins considered undruggable.
The technology is early, but the tipping point is here. These advances are beginning to have concrete effects on drug discovery and development.
CEO Sean McClain expects Absci’s AI-enhanced approach to slash the time and costs needed to bring new molecules to the clinic, from a 5-and-a-half-year process costing up to $100 million to a 2-year process costing $15 million.
“We’re really starting to break the biotech economics,” McClain said. “Instead of investing traditionally $100 million into one drug asset, you can now invest it into five to 10 and get them into the clinic much faster.”
Starting From Scratch
Proteins— there are about 20,000 in the human body — handle much of the heavy lifting required for life. They give cells structure but also send important messages and process signals. Some proteins aid embryonic development or stop cells from turning cancerous. Disease-fighting antibodies are proteins. So are enzymes and many hormones.

Proteins are formed by strings of electrically charged amino acids that attract or repel one another, causing folding that gives proteins distinct shapes. These shapes help determine each protein’s function.
Researchers have long sought to understand how proteins fold into shape in hopes of learning how to build them from scratch. Even as recently as 2017 — the year of a flu-fighting protein design breakthrough — protein design was still fairly inefficient.
“The software just would not work reliably every single time that you’d want it to,” Anand said.
But AI has since taken off. In 2022, the open-access AlphaFold Protein Structure Database, developed by Google DeepMind and the European Bioinformatics Institute, made a trove of more than 200 million protein structure predictions searchable for any researcher. In drug discovery, these protein structures can provide starting points for finding an initial “hit” to optimize.
This “high-quality, large dataset of proteins” is the only one of its kind, said Gregory Bowman, PhD, a professor at the University of Pennsylvania, in Philadelphia. “But it doesn’t mean take AlphaFold, adjust new drug discovery, and presto, get a bunch of drugs.”
A potential snag: “A large fraction of proteins are considered undruggable,” Bowman said. That is, they lack cavities where drugs could bind or those cavities are too subtle for most technologies to find.
Bowman’s lab used computer simulations to hunt down those “cryptic pockets” and then followed up with experiments to confirm their predictions. They built a dataset and used it to train a neural network, called PocketMiner, which uses machine learning to predict where cryptic pockets might lie.

Last year, PocketMiner predicted cryptic pocket locations in nearly three dozen cancer-related protein structures, opening the door for potential new treatments.
“I think these cryptic pockets are in most proteins, which means that they’re relevant to most cancers,” Bowman said.
His lab is collaborating with start-up companies on breast cancer and uveal melanoma, a cancer of the eye. They’re also exploring targets for diseases such as leukemia and pancreatic cancer.
That research can complement de novo design efforts, said Anand. “In my mind, they’re two halves of the same coin. If you have very good prediction of where to bind, then you can design a drug against said pocket.”
And if researchers generate thousands of de novo proteins, a solid predictive model can help determine which design is more likely to work. “There’s a way to weave them together,” Anand said.
Generating Protein Structures and Sequences
Some proteins are more challenging to design than others. Take helical peptides — short, flexible, coil-like chains of amino acids found in very small concentrations in the body. Designing proteins that can bind to such elusive structures has been a struggle. But a team at Baker Lab, part of the Institute for Protein Design at the University of Washington, used two deep learning tools to pull it off.
Their efforts, published last December, could make diagnosing diseases cheaper and faster, and lead to therapeutics that bind to hormones to block their activity. For instance, their designed proteins target parathyroid hormone (PTH), a parathyroid cancer biomarker, and neuropeptide Y, associated with Alzheimer’s disease. Their de novo protein detected PTH by mass spectrometry and enabled protein biosensors responsive to PTH.
“Making antibodies for some of these targets, which are the gold-standard molecules for diagnostics or therapeutics, could take months,” said lead author Susana Vazquez Torres, PhD, a researcher at the Baker Lab. But with AI, researchers can generate thousands of proteins in just one day.

They start with a target helical peptide (such as PTH) surrounded by a random distribution of atoms, like “a cloud of noise.” Next, a diffusion model “de-noises” the atoms, causing them to fold into the shape of a protein. Again, this technology borrows from the AI-powered text-to-image models mentioned earlier, which are “trained” on troves of online images and text.
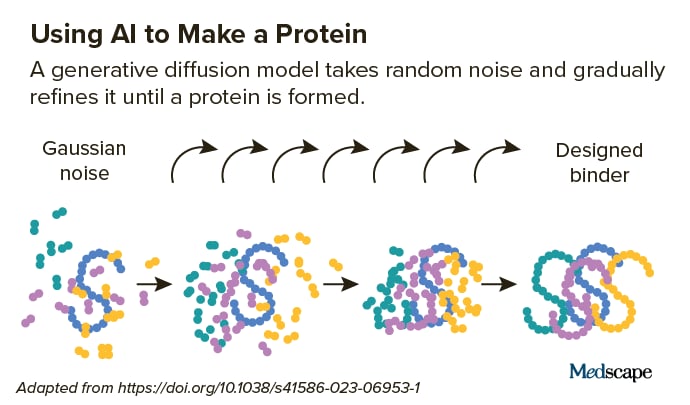
“Diffusion is a very powerful technology because you don’t necessarily need to prespecify the shape of the protein,” said Vazquez Torres.
The Baker Lab trained their diffusion model on images in the Protein Data Bank, a vast archive of 3D protein structure data. The model can quickly generate thousands of different protein backbones of various shapes that fold differently around the target. Next, the lab’s deep learning algorithm, Protein MPNN, generates amino acid sequences that interact with the target. The model then ranks the highest-affinity binders, which are further studied.
Vazquez Torres has been applying the same techniques to design de novo proteins targeting the most toxic components in snake venom. She hopes they can become new treatments for developing regions, where snake bites kill about 100,000 people per year and often result in amputations. De novo proteins are cheaper to produce than current therapeutics and can withstand the regions’ high temperatures, according to Vazquez Torres.
“With protein design, in the span of a week, you can come up with a very good binder. Now, it’s the moment to look for exciting applications where we could have an impact,” she said.
Tackling the Antibody Challenge
The therapeutic antibody market is worth an estimated $247 billion and is expected to grow 14% by 2028. AI is helping fuel that growth.

Antibodies undergo a natural evolution process in the body, gaining efficacy against a virus over a few weeks, according to Brian Hie, PhD, an assistant professor at Stanford University, California. With machine learning, a subset of AI that trains models to continuously learn from data, Hie and his colleagues guide antibody evolution in the lab.
Their protein language models predict which mutations are most likely to occur among all possible mutations to an antibody sequence. The process is much faster than traditional approaches that randomly introduce mutations to vast collections of antibodies and then use protein engineering techniques, such as yeast surface display, to assess binding activity.
“We wanted not to have to screen a million antibodies every time we wanted to evolve an antibody to have higher affinity,” Hie said.
The evolved antibodies can become the basis for new or improved therapeutics. In a preprint published last December, the Stanford researchers used antibodies that had lost efficacy against the Omicron variant of severe acute respiratory syndrome coronavirus 2 (SARS‑CoV‑2) to create new antibodies that are more effective against viral variants. Hie’s lab is also exploring applications in cancer, as “tumors are constantly evolving around chemotherapy or immunotherapy,” Hie said.
Antibodies’ flexible loops have often stymied AI models, but de novo design breakthroughs are ticking up. In a preprint published in March, the Baker Lab showed how an adjusted version of their diffusion model — trained on additional antibody structures — could design antibodies capable of recognizing a cancer drug target and bacterial and viral proteins related to flu and SARS-CoV-2. About one in 100 designs binded to their targets in follow-up lab tests.
Absci is getting closer to delivering an improved antibody to patients with inflammatory bowel disease, thanks to its AI models. Preclinical tests suggest their anti-TL1A antibody would need less frequent dosing and offer higher potency and better clinical safety compared with competitor molecules currently in clinical trials.
“It’s not enough to be able to say you can de novo design an antibody with AI,” said Absci CEO McClain. “The bigger inflection points are going to be once there is clinical proof-of-concept that these models are living up to their promise.”


